如今,深度学习技术发展迅速[3-4],卷积神经网络(Convolutional Neural Network,CNN)以其强大的特征提取能力在图像分析领域取得了重大突破[5-6],基于CNN的U型网络结构(U-Net)[7]在医学图像分割中已被广泛应用.U-Net是一种对称的编码器-解码器结构,该结构在编码器到解码器之间加入了跳跃连接,使得网络更好地融合不同尺度的特征.但由于卷积操作本身仅进行局部运算,很难构建像素间长距离依赖关系,所以U-Net结构仍有很大的改进空间.Li等[8]利用不同空洞率的空洞卷积提取多尺度目标特征,改善了右心室的分割结果,该方法通过引入空洞卷积来增大感受野,但需要结合多个不同空洞率的卷积,增大了计算复杂度.Cheng等[9]将方向场应用到U-Net中,通过方向场的监督来减少相似区域的误分割,但需调整相应的权重系数.罗恺锴等[10]在U-Net结构中引入了通道注意力机制,采用多视角融合的方法提升了脑肿瘤MRI的分割精度,王瑞豪等[11]则结合切片上下文信息,分成多个阶段完成胰腺的分割,上述两种方法虽然取得了较高的精度,但整体流程比较复杂.Yu等[12]将自注意力机制[13]嵌入到U-Net结构中,提高了心脏MRI的分割准确率,但该方法仅将自注意力机制模块嵌入到网络中分辨率较低的特征图后,导致其提升效果有限.
Transformer[14]结构不受限于局部运算,能够建模全局上下文信息,在自然语言处理任务上有着出色的表现.Dosovitskiy 等[15] 提出了ViT(Vision Transformer),首次将Transformer 结构应用于图像分类任务,超越了基于CNN方法的分类精度.Zheng 等[16]结合ViT结构,将语义分割转化为序列形式的预测任务,开辟了语义分割任务的新范式,但ViT结构输出特征图的分辨率低且单一,导致局部信息的丢失.Chen等[17]和李耀仟等[18]均在U-Net的最小特征图后引入Transformer结构,将卷积层提取的特征转换为序列输入到Transformer中,从而捕获全局依赖关系,但其参数量大、计算复杂度较高.Cao等[19]参考Swin Transformer[20],分成多个阶段产生不同尺度的特征,提出了基于编码器-解码器的纯Transformer结构,改善了腹部和心脏图像分割结果,但该结构需要预训练权重才能发挥效果,导致其网络结构不能灵活调节.
针对心脏MRI分割当前面临的技术问题,本文提出了一种全局和局部信息交互的双分支U型网络(UConvTrans),该模型在CNN基础上引入Transformer结构,不仅能提取局部信息特征,还增强了网络提取全局信息特征的能力.此外,本文提出的融合CNN及Transformer结构的模块(Fuse CNN and Transformer Block, FCTB)分别将CNN分支的输出和Transformer分支的输出相互拼接实现特征交互融合,增强了模型表达能力.最终,在MICCAI 2017 ACDC数据集[21]上的大量实验结果表明,和其他基于CNN或者基于Transformer的方法相比,UConvTrans仅在较少的参数下,实现了目标区域的准确分割.
1 方法
1.1 研究基础
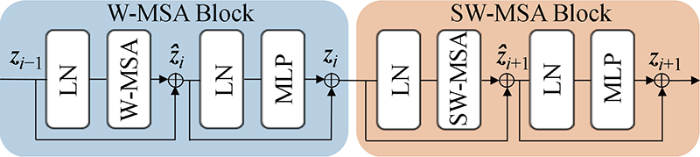
CNN通过与周围像素点运算来提取局部特征,具有平移不变性,但受到卷积核大小与计算资源的限制,导致其建模全局信息能力不足.而Transformer结构得益于其多头自注意力机制强大的全局信息提取能力,可以构建像素间长距离的依赖关系.心脏MRI的心室和心肌区域聚集较近,各类别边界之间比较相似,有效地处理全局信息能够对分割精度提升带来很大帮助.最近,基于Transformer结构的方法在多个视觉任务榜单中已超过基于CNN的方法,Swin Transformer[20]表现尤为出色,其核心模块(Block)如图1所示.图中:zi表示第i个模块经多层感知机(Multi-Layer Perceptron,MLP)和残差连接后输出的特征;
图1
每个Swin Transformer 模块均包含了两个连续的多头自注意力模块,每个模块均由层归一化(Layer Normalization,LN)、多头自注意力机制、残差连接以及MLP组成,多层感知机由两层线性层及高斯误差线性单元(Gaussian Error Linear Unit,GELU)组成.其中第1个模块应用了W-MSA,第2个模块为SW-MSA.整体计算过程如下:
在计算多头(head)自注意力机制时,每个head的计算方式如下:
式中:Q, K, V∈
Swin Transformer中基于窗口的自注意力机制降低了原本自注意力机制的计算复杂度,整体框架借鉴CNN基网络[22]中的层级结构分成多个阶段来获取不同尺度的特征.而医学图像数据集规模较小,没有通用的预训练权重,且目标类别相对自然图像而言也较少,分割精度要求更高,较大的模型会导致过拟合.因此与原本的Swin Transformer不同,本文提出的UConvTrans的核心模块FCTB以CNN和Transformer相互融合的方式进行特征交互,FCTB中的Transformer结构无需预训练权重,可以灵活调整网络结构,并且FCTB中的Transformer结构结合CNN来补充局部位置信息,无需位置编码.
1.2 UConvTrans 总体结构
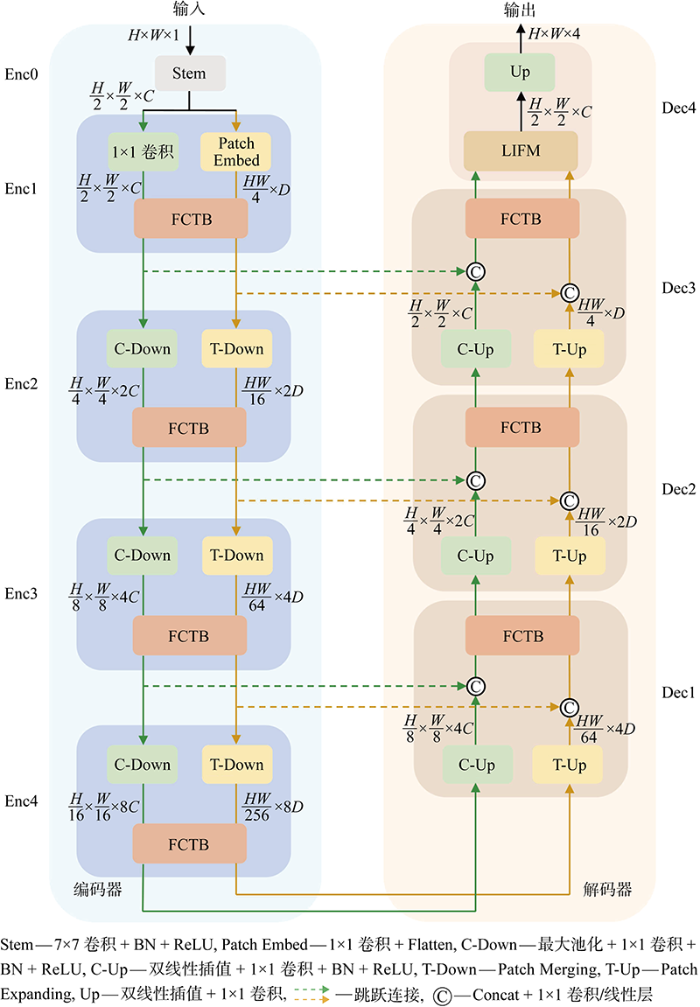
为了增强网络上下文信息感知能力以及保留丰富的细节信息,针对心脏MRI数据特点,提出了一种CNN和Transformer相互融合的双分支分割网络框架,其整体结构基于编码器-解码器的形式,如图2所示.图中:C表示模型的基础特征图通道数;D表示模型的基础序列特征维度数;H和W代表输入图像的高度和宽度;LIFM为最终的信息整合模块(Last Information Fusion Module,LIFM).
图2
在编码器中,主要分为5个阶段,即图2中的Enc0到Enc4.首先,大小为H×W×1(输入图像通道数为1)的图像经过Stem模块来提取初始特征,Stem由大小为7×7,步长为2的卷积、批标准化(Batch Normalization, BN)和线性修正单元(Rectified Linear Unit,ReLU)组成,其输出特征图的大小为H/2×W/2×C.接着在Enc1阶段,Stem模块的输出在CNN分支经过1×1卷积后输入到FCTB中,在Transformer分支经过Patch Embed后进入FCTB中,Patch Embed具体过程为先经过1×1卷积将特征图映射为H/2×W/2×D大小,然后经Flatten操作后展开为序列的形式,序列特征大小为HW/4×D.然后在Enc2阶段,卷积分支通过最大池化层将分辨率大小为H/2×W/2的特征图降低为H/4×W/4,并通过1×1卷积将通道数C增大为2C,得到大小为H/4×W/4×2C的特征图.Transformer分支通过Patch Merging[20]将大小为HW/4×D 的序列特征转换为HW/16×2D,之后一同输入到FCTB中,后续Enc3到Enc4这两个阶段处理过程与Enc2阶段类似.编码器分为多个阶段来提取浅层的空间信息和深层的语义信息,其CNN分支为Transformer分支提供局部特征和位置信息,Transformer分支为CNN分支提供全局上下文信息.
在编码器提取深层次特征后,UConvTrans通过解码器来降低特征图和序列特征的维度并恢复原始输入尺寸,解码器分为4个阶段,即图中的Dec1到Dec4.在Dec1阶段中,CNN分支通过双线性插值将分辨率大小为H/16×W/16的特征图增大为H/8×W/8,并通过1×1卷积将通道数由8C减少为4C,得到大小为H/8×W/8×4C的特征图.然后该特征图与编码器Enc3阶段的卷积分支输出的特征图进行跳跃连接,在通道维度上拼接(Concat)后得到大小为H/8×W/8×8C的特征图,经1×1卷积将通道数降低到4C后得到大小为H/8×W/8×4C的特征图.Transformer分支通过Patch Expanding[19]将大小为HW/256×8D的序列特征转化为HW/64×4D,然后与编码器Enc3阶段的Transformer分支输出的序列特征进行跳跃连接,经Concat后得到HW/64×8D的序列特征,之后通过线性层将维度数降低为4D,得到大小为HW/64×4D的序列特征,然后两分支将各自处理的结果输入到FCTB中.Dec2,Dec3的处理过程与Dec1类似,Dec3阶段两分支的输出通过LIFM后,再上采样到原图大小,经1×1卷积得到最终的分割预测图,其大小为H×W×4(4为类别数,包含背景).为了减少编码器下采样时丢失的信息,解码器每次上采样后的特征和编码器提取的特征通过跳跃连接进行融合,进而能够改善心脏MRI多尺度目标和心室及心肌轮廓细节的分割.
1.3 融合CNN及Transformer结构的模块
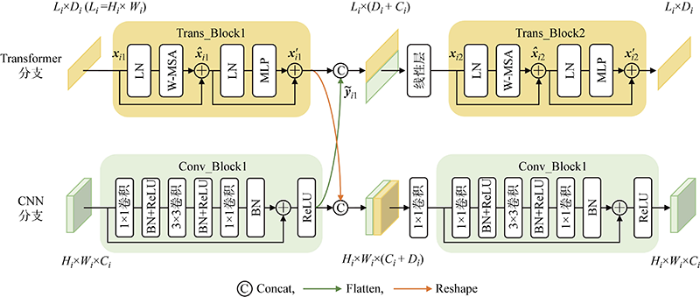
本文提出的融合CNN及Transformer结构的FCTB模块能够利用CNN和Transformer的各自优势分别提取局部和全局特征,并通过交互融合的方式,既构建了上下文依赖关系,又丰富了局部细节信息,增强了网络提取特征的能力.FCTB是编码器和解码器各个阶段的核心模块,其详细结构图如图3所示,由两个分支构成,一个为Transformer分支,另一个为CNN分支.
图3
1.3.1 FCTB中的Transformer分支
Transformer分支主要由两个连续的多头自注意力模块(Trans_Block)组成,为了减少计算复杂度,本文将Transformer原生的多头自注意力机制改进为W-MSA[20],每个模块由LN、W-MSA、残差连接以及MLP组成.假设在第i个阶段,Transformer分支中第1个模块(Tran_Block1)的输入为xi1、经W-MSA和残差连接输出的变量为
为了补充位置信息和增强局部特征表达,Trans_Block1的输出与Conv_Block1的输出进行融合.其中,Trans_Block1的输入序列特征大小为Li×Di,Li为在第i个阶段的序列特征长度,Di为第i个阶段的序列特征维度数,Conv_Block1的输出特征图大小为Hi×Wi×Ci,Hi和Wi为在第i个阶段的特征图的高与宽,Ci为第i个阶段特征图的通道数,并且Li=Hi×Wi.首先将Conv_Block1的输出展开成序列特征的形式,展开后大小为Li×Ci,即图3中的Flatten操作,之后与Trans_Block1的输出在特征维度上进行拼接,此时大小为Li×(Di+Ci),接着经一个线性层将维度数Di+Ci降低到Di,再输入到Trans_Block2中进行处理,Trans_Block2的计算过程与Trans_Block1一致,最终输出序列特征大小为Li×Di.
通过引入Transformer结构,网络增强了提取全局信息特征的能力,并且由于Transfomer分支在计算时得到了CNN分支提供的位置信息和局部特征,所以FCTB舍弃了Transformer原本的位置编码,通过融合CNN提取的特征来获得位置信息,从而避免了固定输入尺寸的限制.
1.3.2 FCTB中的CNN分支
CNN分支主要由两个连续的卷积残差瓶颈模块[22](Conv_Block)组成,其处理过程与Transformer分支类似,该模块包括1×1卷积、3×3卷积、BN和ReLU激活函数,在输入和瓶颈模块的输出之间通过残差连接来加速模型收敛.
为了弥补CNN建模全局信息能力的不足,将Conv_Block1的输出与Trans_Block1的输出进行融合.其中,Conv_Block1输出的特征图大小为Hi×Wi×Ci,Trans_Block1输出的序列特征大小为Li×Di.首先将Trans_Block1的输出进行转换,转换后得到大小为Hi×Wi×Di的特征图,即图3中的Reshape操作,然后和Conv_Block1的输出在通道维度上进行拼接,此时特征图大小为Hi×Wi×(Ci+Di),最终经过一个1×1的卷积将维度数Ci+Di降低到Ci后,再输入到Conv_Block2中,最终卷积分支输出特征图的大小为Hi×Wi×Ci.CNN分支能够提取局部特征,而融合了Transformer分支的输出后,增强了模型建模上下文信息的能力.
1.4 最终的信息融合模块
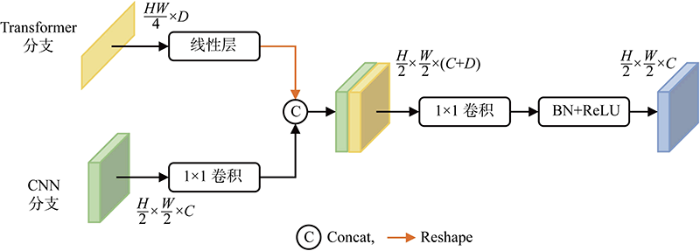
最终的信息融合模块LIFM用于融合Transformer分支和CNN分支的最终输出,其结构如图4所示.解码器中两分支的输出通过融合模块进行融合,Transformer分支的输出首先经过线性层,之后进行Reshape操作,CNN分支的输出经过1×1卷积后,与Transformer分支Reshape后的特征在通道维度上进行拼接,之后经过1×1卷积降低维度.LIFM输出的特征图双线性插值到原图大小,经1×1类别映射卷积得到分割结果.
图4
1.5 模型参数配置
该模型无需预训练权重,模型结构参数可以灵活调整,本文的模型参数有以下两种配置:① 轻量化模型为C=32,D=32,该配置下的模型参数量仅为3.65×106;② 高精度模型为C=32,D=64,该配置下的模型参数量为1.059×107.需要指出的是,以上两种模型参数配置相较于经典的CNN[22]和最近提出的视觉Transformer[20]网络结构,基础特征图维度数和序列特征维度数都要更低,因此整体模型的参数量较少,模型的运行效率更高.此外,Transformer结构其他的参数配置为:W-MSA[20]中Window尺寸为8,MLP中线性层的维度变化率为2,编码器中Enc1到Enc4四个阶段中W-MSA的head数分别为2,4,8,16,解码器Dec1到Dec3三个阶段中W-MSA中的head数分别为8,4,2.
1.6 损失函数
本文使用的损失函数为Soft Dice Loss,具体计算公式如下:
式中:M为类别个数;N为像素点总个数;模型最终输出经过Softmax函数后得到Pi, j,Pi, j为第i个像素点被分类为第j个类别的概率;Ti,j为经 onehot编码后的标签,表示第i个像素点属于第j个类别.
2 实验与分析
2.1 数据介绍及处理
本文实验使用MICCAI 2017 ACDC数据集,该数据集包含150个患者的短轴心脏MRI,提供了在舒张末期和收缩末期时刻的分割标签,由医学专家手动标注完成.该数据集由两个不同磁性强度的MRI扫描仪进行采集,其切片层间距离为5 mm或者10 mm,空间分辨率为1.37 ~1.68 mm2/像素.其中训练集包含100个公布分割标签的患者数据,包含4个区域:背景、右心室(Right Ventricle,RV)、心肌(Myocardium,Myo)、和左心室(Left Ventricle,LV).另外测试集包含50个未公布分割标签的患者数据,需要在官网上提交分割结果返回得分.此外,根据其疾病类型,150个MRI数据被均匀划分为5组,疾病种类分别为正常、心力衰竭伴梗死、扩张型心肌病、肥厚型心肌病和右心室异常.每个患者的MRI空间尺寸及切片数变化不一,整体MRI的高度范围为154~428 像素,平均高度为219.72 像素,宽度范围为154~512 像素,平均宽度为243.15 像素,切片数范围为6~21,平均切片数为10.单个病例的心脏MRI示意图如图5所示.由于该数据集MRI层与层的间隔较大、切片数量较少,按照以往的工作经验[23-24],本文将每个患者在舒张末期以及收缩末期的心脏MRI在切片维度进行切片,将其处理成二维形式,最终将训练集切片成 1 902 张图像.
图5
由于在官网测试集上的提交次数有限,本文将原训练集划分为训练集和验证集,消融实验在验证集上进行验证,最终在官方测试集上评估模型性能.为了保证每种疾病的患者图像均参与训练,分别在5种疾病类型中随机选取16个患者数据为训练集,其余4个为验证集,将有标签的100个患者数据划分为含80例患者的训练集和20例患者的验证集.为了防止过拟合以及扩充训练数据,对每张切片采取以下在线数据增强方式:随机水平翻转、随机角度旋转、随机多尺度变化以及随机裁剪.对于尺寸小于256 像素的切片进行零填充,最终输入图像尺寸为256 像素×256 像素,之后对每张图像进行标准化处理.
2.2 评价指标
为了评估模型性能,使用了Dice系数(Dice Similarity Coefficient,DSC)作为评价标准.Dice系数衡量分割标签和预测结果的相似程度,数值范围为0~1,0表示相似程度最小,1表示相似程度最大.DSC计算公式如下:
式中:NTP表示预测的像素点被正确分类为目标类别的数量;NFP表示预测的像素点被错误分类为目标类别的数量;NFN表示预测的像素点被错误分类为非目标类别的数量.
2.3 实验环境和参数
实验在i9-9820X CPU、两块NVIDIA RTX 2080 Ti GPU的设备上进行,操作系统为Ubuntu 20.04,并在PyTorch框架下实现网络模型.实验的总轮数设置为 1000 轮,批数大小设为16,初始的学习率设为0.01,使用warming up预热1轮后采取poly学习率衰减策略,衰减率为0.9.采用随机梯度下降(Stochastic Gradient Descent, SGD)作为模型的优化器,动量为0.9,权重衰减项设置为 1×10-4.
2.4 消融实验
2.4.1 核心模块的有效性
为了验证FCTB的有效性,本文对比了UConvTrans中的CNN分支、Transformer分支以及两个分支是否融合的结果,实验结果如表1所示.
表1 FCTB消融实验结果
Tab.1
| 方法 | Fuse Trans to Conv | Fuse Conv to Trans | DSC /% | |||
|---|---|---|---|---|---|---|
| 平均 | RV | Myo | LV | |||
| Only Trans | — | — | 83.75 | 80.75 | 82.48 | 88.02 |
| Only Conv | — | — | 87.60 | 86.64 | 86.17 | 89.98 |
| Trans+Conv | × | × | 88.61 | 86.70 | 87.72 | 91.40 |
| Trans+Conv | × | √ | 88.76 | 87.52 | 87.06 | 91.69 |
| Trans+Conv | √ | × | 89.25 | 87.08 | 88.31 | 92.38 |
| Trans+Conv | √ | √ | 89.38 | 87.12 | 88.44 | 92.57 |
首先对比前3组实验,实验的模型分别为仅有Transformer分支(Only Trans)、仅有CNN分支(Only Conv)以及有两分支但FCTB中不进行融合(Trans+Conv)的结构.实验表明,仅有Transformer分支的网络平均Dice系数低于仅有CNN分支的网络,而结合两分支后平均Dice系数达到了88.61%,相比于单分支网络有了较大的提升.这说明本文模型当仅有Transformer分支时,在该实验数据中表现较差,而结合CNN和Transformer两种结构可以提高分割精度.
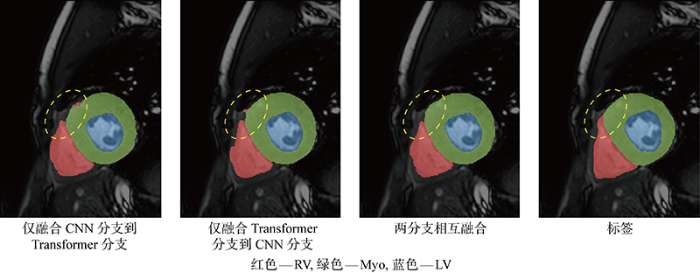
然后,后3组实验分别是在有了两分支的基础上,在FCTB中仅融合CNN分支提取的特征到Transformer分支中的结构(Fuse Conv to Trans)、仅融合Transformer分支提取的特征到CNN分支中的结构(Fuse Trans to Conv)以及二者相互融合完整的FCTB结构.相比于未融合的双分支结构,融合CNN分支提取的特征到Transformer分支中平均提升了0.15%,融合Transformer分支提取的特征到CNN分支中平均提升了0.64%,Transformer分支和卷积分支相互融合平均提升了0.77%,上述3种结构的分割结果如图6所示,二分支相互融合的方式得到的分割结果更加细腻.结果表明CNN分支得到Transformer分支提取的全局信息特征能够有效提升分割精度,当Transformer分支得到CNN分支补充的位置及局部特征后也会有轻微的提升, 而本文提出的FCTB模块也能进一步提升两个分支的融合效果.
图6
2.4.2 模型参数的有效性
为了验证模型参数配置的有效性,此部分实验对比了在不同参数情况下对分割性能的影响.对于CNN分支主要通过基础特征图通道数C控制其参数量,对于Transformer分支通过基础序列特征维度数D控制其参数量,结果如表2所示.相对于C=32,D=32,当增加Transformer分支的参数时(C=32, D=64),RV区域的得分提升了近1%,其余部分变化不大,而当CNN分支的参数增加时(C=64, D=32),RV区域的得分提升了0.37%,但其余区域均有所下降,同时增加CNN和Transformer的参数量(C=64, D=64),并没有提升整体的分割性能.结果表明:相比于CNN分支的参数量、计算量,整体的参数量和计算量受Transformer分支的影响更大,增大Transformer分支的参数能够带来一定的提升,但其参数量、计算量也会成倍增加,而采用第一组实验(C=32, D=32)的配置能更好地平衡效率和精度.
表2 模型参数的消融实验结果
Tab.2
| 通道数 | 维度数 | DSC/% | 参数量×10-6 | 计算量×10-9 | |||
|---|---|---|---|---|---|---|---|
| 平均 | RV | Myo | LV | ||||
| 32 | 32 | 89.38 | 87.12 | 88.44 | 92.57 | 3.65 | 5.03 |
| 32 | 64 | 89.60 | 88.08 | 88.30 | 92.41 | 10.59 | 12.74 |
| 64 | 32 | 88.97 | 87.49 | 87.81 | 91.60 | 7.39 | 10.81 |
| 64 | 64 | 89.30 | 87.80 | 88.17 | 91.92 | 14.54 | 18.79 |
2.5 对比实验
为说明本网络模型在心脏MRI分割方面的优势,首先在本地划分的验证集上与经典医学图像分割算法以及最近提出的基于Transformer的医学图像分割算法进行比较,实验结果如表3所示.在C=32,D=32的配置下,和U-Net、Attention U-Net以CNN为基础的网络相比,本文网络结构的参数量和计算量约为U-Net、Attention U-Net的10%和8%,但平均Dice系数分别提升了1.13%、0.86%.和基于Transformer的两个网络相比,本文的Transformer结构在原结构参数的基础上进行了修改,而得益于信息交互融合的设计,本文的Transformer结构在不需要预训练权重的情况下,同样能够有效提升模型分割精度.本模型在C=32,D=32的参数配置下,其参数量、计算量却仅为TransUnet的3.47%、13.04%,但比TransUnet的平均得分仅低了0.09%,比SwinUnet平均提升了0.12%.而且本模型在C=32,D=64的参数配置下,能够取得最高的平均Dice系数.相比于表3中的其他模型,本模型设定的特征图通道数或者序列特征维度数较小,因此整体模型的参数量也较小,并且能够保持模型的精度.本模型以CNN和Transformer相互融合的方式来搭建整体网络,能够结合CNN和Transformer的优点,从而增强模型的特征提取能力,并且无需预训练权重,模型结构参数可以灵活调整.
表3 本文的方法和其他方法在验证集上的比较结果
Tab.3
| 方法 | DSC/% | 参数量×10-6 | 计算量×10-9 | |||
|---|---|---|---|---|---|---|
| 平均 | RV | Myo | LV | |||
| U-Net[7] | 88.25 | 86.91 | 87.17 | 90.65 | 34.53 | 65.55 |
| Attention U-Net[25] | 88.52 | 86.78 | 86.93 | 91.84 | 37.88 | 66.62 |
| SwinUNet[19] | 89.26 | 86.62 | 88.72 | 92.44 | 27.17 | 6.14 |
| TransUNet[17] | 89.47 | 87.04 | 88.51 | 92.85 | 105.32 | 38.57 |
| UConvTrans (C=32,D=32) | 89.38 | 87.12 | 88.44 | 92.57 | 3.65 | 5.03 |
| UConvTrans (C=32,D=64) | 89.60 | 88.08 | 88.30 | 92.41 | 10.59 | 12.74 |
表4 在MICCAI 2017 ACDC 测试集上的比较结果
Tab.4
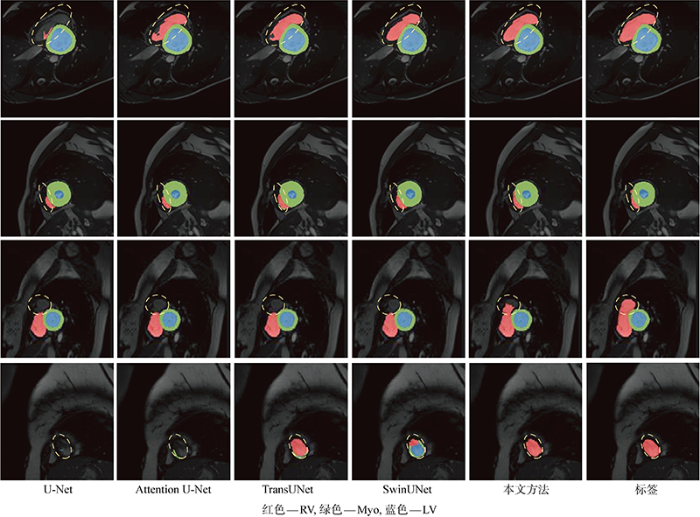
最后,图7展示了多个模型的分割结果及标签可视化对比,每一行是在本地划分的验证集中挑选的病例切片,每一列是不同模型的可视化结果,本文模型采用了C=32,D=64的参数配置.前两行的可视化结果可以观察到,和基于CNN的方法相比,本方法提取全局特征能力更强,得到的分割结果更加准确,和基于Transformer的方法相比,本方法可以有效地保留细节信息,从而使目标区域的轮廓更为平滑.第3行的结果可以看出,对于形状变化不一的RV区域,本方法改善了该区域欠分割的问题.在最后一行结果中,本文方法和真实标签更为接近,没有出现类别区域误分割的问题.以上实验说明了本方法可以提高RV这类困难区域的分割精度,并且模型判别能力强,能够准确识别心脏MRI中复杂多变的背景和目标区域.
图7
图7
不同模型的分割结果可视化
Fig.7
Visual comparison of cardiac segmentation results of different methods
3 结论
针对心脏MRI分割任务,本文的工作主要有:
(1) 提出了一种全局和局部信息交互的双分支分割网络,该结构通过CNN分支提取局部特征,Transformer分支建模全局上下文信息,能更好地识别轮廓细节并且抑制背景干扰,从而有效地应对了心脏MRI分割的难点.
(2) 设计了一个融合模块,有效融合了CNN提取的局部特征和Transformer提取的全局特征,提高了网络的判别能力,并且本文模型中的Transformer结构不需要在大规模数据上进行预训练.
(3) 在公开数据集MICCAI 2017 ACDC上验证了该方法的有效性,模型的参数量、计算量较少,更好地平衡了精度和效率,而且在官方测试集中的心肌和左心室区域取得了到目前为止最高的Dice得分.虽然本文提出的模型能准确分割心脏MRI的目标区域,但该模型是二维网络,更适用于需要对原三维图像切片成二维图像的心脏MRI.在后续研究中,将对该模型进一步改进及优化,使其更有效地应用到切片信息更加丰富的三维医学影像分割中.
参考文献
Deep learning for cardiac image segmentation: A review
[J].
DOI:10.3389/fcvm.2020.00025
PMID:32195270
[本文引用: 1]

Deep learning has become the most widely used approach for cardiac image segmentation in recent years. In this paper, we provide a review of over 100 cardiac image segmentation papers using deep learning, which covers common imaging modalities including magnetic resonance imaging (MRI), computed tomography (CT), and ultrasound and major anatomical structures of interest (ventricles, atria, and vessels). In addition, a summary of publicly available cardiac image datasets and code repositories are included to provide a base for encouraging reproducible research. Finally, we discuss the challenges and limitations with current deep learning-based approaches (scarcity of labels, model generalizability across different domains, interpretability) and suggest potential directions for future research.Copyright © 2020 Chen, Qin, Qiu, Tarroni, Duan, Bai and Rueckert.
Seg-CapNet: 心脏MRI图像分割神经网络模型
[J].
Seg-CapNet: Neural network model for the cardiac MRI segmentation
[J].
深度神经网络模型压缩综述
[J].
A survey of model compression for deep neural networks
[J].
医学图像分析深度学习方法研究与挑战
[J].
Deep learning in medical image analysis and its challenges
[J].
低剂量CT图像去噪的改进型残差编解码网络
[J].
Improved residual encoder-decoder network for low-dose CT image denoising
[J].
基于距离置信度分数的多模态融合分类网络
[J].
Multimodal fusion classification network based on distance confidence score
[J].
U-Net: Convolutional networks for biomedical image segmentation
[C]
Dilated-inception net: Multi-scale feature aggregation for cardiac right ventricle segmentation
[J].DOI:10.1109/TBME.10 URL [本文引用: 1]
Learning directional feature maps for cardiac MRI segmentation
[C]
引入注意力机制和多视角融合的脑肿瘤MR图像U-Net分割模型
[J].
U-Net segmentation model of brain tumor MR image based on attention mechanism and multi-view fusion
[J].
结合切片上下文信息的多阶段胰腺定位与分割
[J].
DOI:10.12263/DZXB.20200101
[本文引用: 1]

当前基于深度学习的胰腺分割主要存在以下问题:(1)胰腺的解剖特异性导致深度网络模型容易受到复杂多变背景的干扰;(2)传统两阶段分割方法在粗分割阶段将整张CT图像作为输入,导致依赖粗分割结果得到的定位不够准确;(3)传统两阶段分割方法忽略了切片间的上下文信息,限制了定位和后续分割结果的提升.针对上述问题,本文提出了结合切片上下文信息的多阶段胰腺定位与分割方法.第一阶段利用解剖先验定位粗略缩小输入区域;第二阶段先使用所设计的DASU-Net进行粗略分割,接着利用切片上下文信息优化分割结果;第三阶段使用单张切片定位进一步减少不相关背景,并使用DASU-Net完成精细分割.实验结果表明,本文所提方法能够有效提高胰腺分割的准确率.
Multi-stage pancreas localization and segmentation combined with slices context information
[J].
DOI:10.12263/DZXB.20200101
[本文引用: 1]

Current deep learning-based pancreas segmentation mainly has the following problems:The anatomical specificity of the pancreas makes the deep network model easily disturbed by complex background;in the traditional two-stage segmentation method,the input of the coarse segmentation is the entire CT image,which leads to inaccurate localization based on the segmentation results;the traditional two-stage segmentation ignores the context information between adjacent slices,which limits the localization and subsequent segmentation results.In order to solve the problems above,a multi-stage pancreas localization and segmentation method combined with slices context information is proposed.In the first stage,anatomical prior locating is used to roughly shrink the input area;in the second stage,the proposed DASU-Net is used for coarse segmentation,and then the segmentation results are optimized with slices context information;last stage,single slice locating is used to further shrink irrelevant background,and then fine segmentation is completed by DASU-Net.The experimental results show that the proposed method can effectively improve the accuracy of pancreas segmentation.
Dual attention U-Net for multi-sequence cardiac MR images segmentation[C]//Myocardial Pathology Segmentation Combining Multi-Sequence CMR Challenge
Non-local neural networks[C]//Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition
Attention is all you need[C]//Advances in Neural Information Processing Systems
An image is worth 16×16 words: Transformers for image recognition at scale[C]//International Conference on Learning Representations
Rethinking semantic segmentation from a sequence-to-sequence perspective with transformers[C]//2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition
TransUNet: Transformers make strong encoders for medical image segmentation
[EB/OL]. (
面向手术器械语义分割的半监督时空Transformer 网络
[J].
Semi-supervised spatiotemporal Transformer networks for semantic segmentation of surgical instrument
[J].
Swin-Unet: Unet-like pure Transformer for medical image segmentation
[EB/OL]. (
Swin Transformer: Hierarchical vision Transformer using shifted windows[C]//Proceedings of the IEEE/CVF International Conference on Computer Vision
Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: Is the problem solved?
[J].
DOI:10.1109/TMI.2018.2837502
PMID:29994302
[本文引用: 2]

Delineation of the left ventricular cavity, myocardium, and right ventricle from cardiac magnetic resonance images (multi-slice 2-D cine MRI) is a common clinical task to establish diagnosis. The automation of the corresponding tasks has thus been the subject of intense research over the past decades. In this paper, we introduce the "Automatic Cardiac Diagnosis Challenge" dataset (ACDC), the largest publicly available and fully annotated dataset for the purpose of cardiac MRI (CMR) assessment. The dataset contains data from 150 multi-equipments CMRI recordings with reference measurements and classification from two medical experts. The overarching objective of this paper is to measure how far state-of-the-art deep learning methods can go at assessing CMRI, i.e., segmenting the myocardium and the two ventricles as well as classifying pathologies. In the wake of the 2017 MICCAI-ACDC challenge, we report results from deep learning methods provided by nine research groups for the segmentation task and four groups for the classification task. Results show that the best methods faithfully reproduce the expert analysis, leading to a mean value of 0.97 correlation score for the automatic extraction of clinical indices and an accuracy of 0.96 for automatic diagnosis. These results clearly open the door to highly accurate and fully automatic analysis of cardiac CMRI. We also identify scenarios for which deep learning methods are still failing. Both the dataset and detailed results are publicly available online, while the platform will remain open for new submissions.
Deep residual learning for image recognition[C]//Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition
An exploration of 2D and 3D deep learning techniques for cardiac MR image segmentation[C]//International Workshop on Statistical Atlases and Computational Models of the Heart
Fully convolutional multi-scale residual DenseNets for cardiac segmentation and automated cardiac diagnosis using ensemble of classifiers
[J].
DOI:S1361-8415(18)30848-X
PMID:30390512
[本文引用: 1]

Deep fully convolutional neural network (FCN) based architectures have shown great potential in medical image segmentation. However, such architectures usually have millions of parameters and inadequate number of training samples leading to over-fitting and poor generalization. In this paper, we present a novel DenseNet based FCN architecture for cardiac segmentation which is parameter and memory efficient. We propose a novel up-sampling path which incorporates long skip and short-cut connections to overcome the feature map explosion in conventional FCN based architectures. In order to process the input images at multiple scales and view points simultaneously, we propose to incorporate Inception module's parallel structures. We propose a novel dual loss function whose weighting scheme allows to combine advantages of cross-entropy and Dice loss leading to qualitative improvements in segmentation. We demonstrate computational efficacy of incorporating conventional computer vision techniques for region of interest detection in an end-to-end deep learning based segmentation framework. From the segmentation maps we extract clinically relevant cardiac parameters and hand-craft features which reflect the clinical diagnostic analysis and train an ensemble system for cardiac disease classification. We validate our proposed network architecture on three publicly available datasets, namely: (i) Automated Cardiac Diagnosis Challenge (ACDC-2017), (ii) Left Ventricular segmentation challenge (LV-2011), (iii) 2015 Kaggle Data Science Bowl cardiac challenge data. Our approach in ACDC-2017 challenge stood second place for segmentation and first place in automated cardiac disease diagnosis tasks with an accuracy of 100% on a limited testing set (n=50). In the LV-2011 challenge our approach attained 0.74 Jaccard index, which is so far the highest published result in fully automated algorithms. In the Kaggle challenge our approach for LV volume gave a Continuous Ranked Probability Score (CRPS) of 0.0127, which would have placed us tenth in the original challenge. Our approach combined both cardiac segmentation and disease diagnosis into a fully automated framework which is computationally efficient and hence has the potential to be incorporated in computer-aided diagnosis (CAD) tools for clinical application.Copyright © 2018 Elsevier B.V. All rights reserved.
Attention U-Net: Learning where to look for the pancreas
[EB/OL]. (
Automatic cardiac disease assessment on cine-MRI via time-series segmentation and domain specific features[C]//International Workshop on Statistical Atlases and Computational Models of the Heart
.
Cardiac MRI segmentation with a dilated CNN incorporating domain-specific constraints
[J].DOI:10.1109/JSTSP.4200690 URL [本文引用: 1]
Learning with context feedback loop for robust medical image segmentation
[J].
DOI:10.1109/TMI.2021.3060497
PMID:33606627
[本文引用: 1]

Deep learning has successfully been leveraged for medical image segmentation. It employs convolutional neural networks (CNN) to learn distinctive image features from a defined pixel-wise objective function. However, this approach can lead to less output pixel interdependence producing incomplete and unrealistic segmentation results. In this paper, we present a fully automatic deep learning method for robust medical image segmentation by formulating the segmentation problem as a recurrent framework using two systems. The first one is a forward system of an encoder-decoder CNN that predicts the segmentation result from the input image. The predicted probabilistic output of the forward system is then encoded by a fully convolutional network (FCN)-based context feedback system. The encoded feature space of the FCN is then integrated back into the forward system's feed-forward learning process. Using the FCN-based context feedback loop allows the forward system to learn and extract more high-level image features and fix previous mistakes, thereby improving prediction accuracy over time. Experimental results, performed on four different clinical datasets, demonstrate our method's potential application for single and multi-structure medical image segmentation by outperforming the state of the art methods. With the feedback loop, deep learning methods can now produce results that are both anatomically plausible and robust to low contrast images. Therefore, formulating image segmentation as a recurrent framework of two interconnected networks via context feedback loop can be a potential method for robust and efficient medical image analysis.
GridNet with automatic shape prior registration for automatic MRI cardiac segmentation[C]//International Workshop on Statistical Atlases and Computational Models of the Heart